Tumor detection in medical imaging using neural networks
Introduction
In the field of medical imaging, the preprocessing of radiographic images is essential in order to effectively detect cancerous tumours. It also involves locating these tumors and estimating their sizes. In this way, doctors can use this information to diagnose the disease but also monitor the progress of treatment.
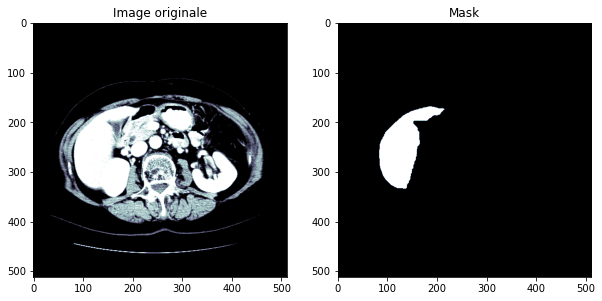
In this project, we will seek to detect cancerous tumors in the liver. We will use open access data that have been annotated by experts (radiologists and oncologists): the tumors are therefore known and localized. The figure below gives an idea of the images that will be used.
Database
The database contains 131 original X-ray images and their corresponding mask images, or 232 images in total, in NII format. The database can be found in the following links part1 and part2.
Study of tumors
We will gather the 232 images in a dataframe containing for each of the 131 original images its source file, its name, the source file of its "mask" image and the name of the latter.
After studying the nii images, we managed to understand our data. Indeed, these are 3D images of dimensions 512 x 512 x n, with n changing from one image to another.
We add two columns to the dataframe: one that shows whether the image contains a tumor or not and the other displays its percentage.
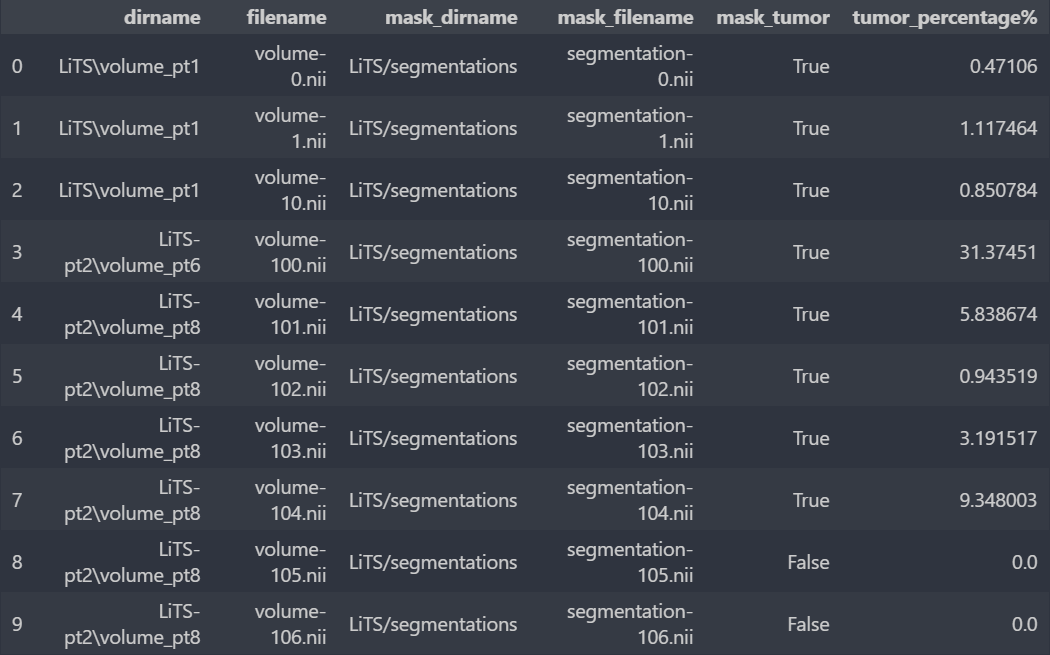
Image conversion
We chose to use the U-Net neural network, a network designed and applied for the first time in 2015 by Oral Ronneberger for the segmentation of medical images.
In addition to having the correct format for U-Net, having jpg or png images will be very useful since our database is very large (49.9 GB), so the manipulation of x-ray images in the NII format takes a lot of time and memory. With this in mind, we are going to transform the 3D images from NII format into 2D images of regular formats: a 3D image of dimensions 512x512x n will be transformed into n images 512x512.
The original images will be transformed into images in jpg format after having preprocessed them thanks to the functions of the fastai library. As for the "mask" images, we will use the png format. This choice is due to the fact that the "labels" 1,2 and 3 contained in an NII "mask" image are lost if this image is transformed into a jpg image, unlike the png format which retains the "labels" well.
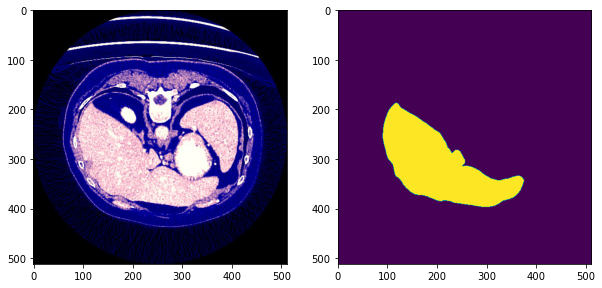
After preprocessing we obtain images of the form:
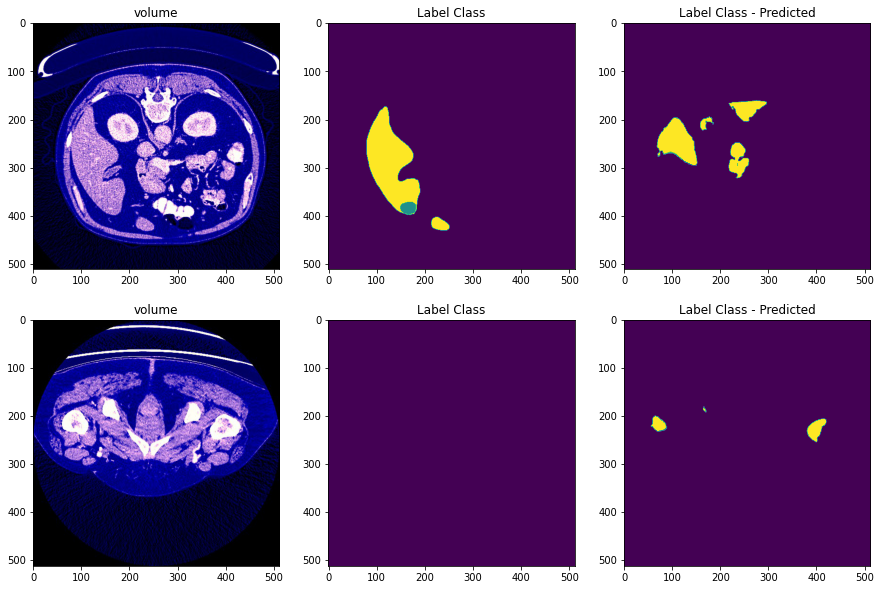
- The first column is the processed image
- The second column is the the mask image
- The third column is the predicted mask of the image
Implementing U-Net
The implementation, training and prediction part is detailed in the report of another team
- After training the images, we obtain the following predicted images